Metal A and Metal B Sites of Nuclear RNA Polymerases Pol IV and Pol V Are Required for siRNA-Dependent DNA Methylation and Gene Silencing | PLOS ONE
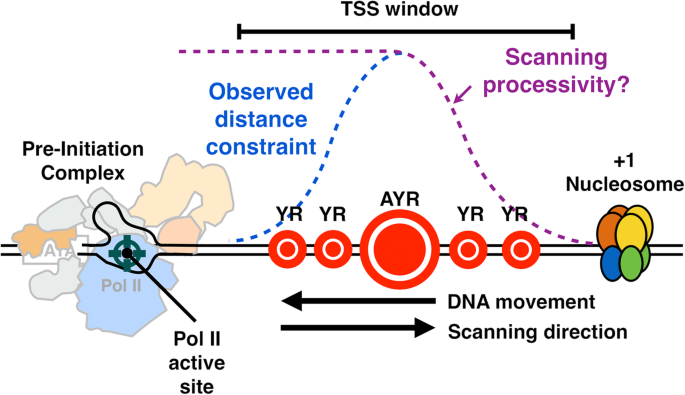
Universal promoter scanning by Pol II during transcription initiation in Saccharomyces cerevisiae | Genome Biology | Full Text

X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct | PNAS

Coordination of RNA Polymerase II Pausing and 3′ End Processing Factor Recruitment with Alternative Polyadenylation | Molecular and Cellular Biology

CUT&RUN detects distinct DNA footprints of RNA polymerase II near the transcription start sites | bioRxiv
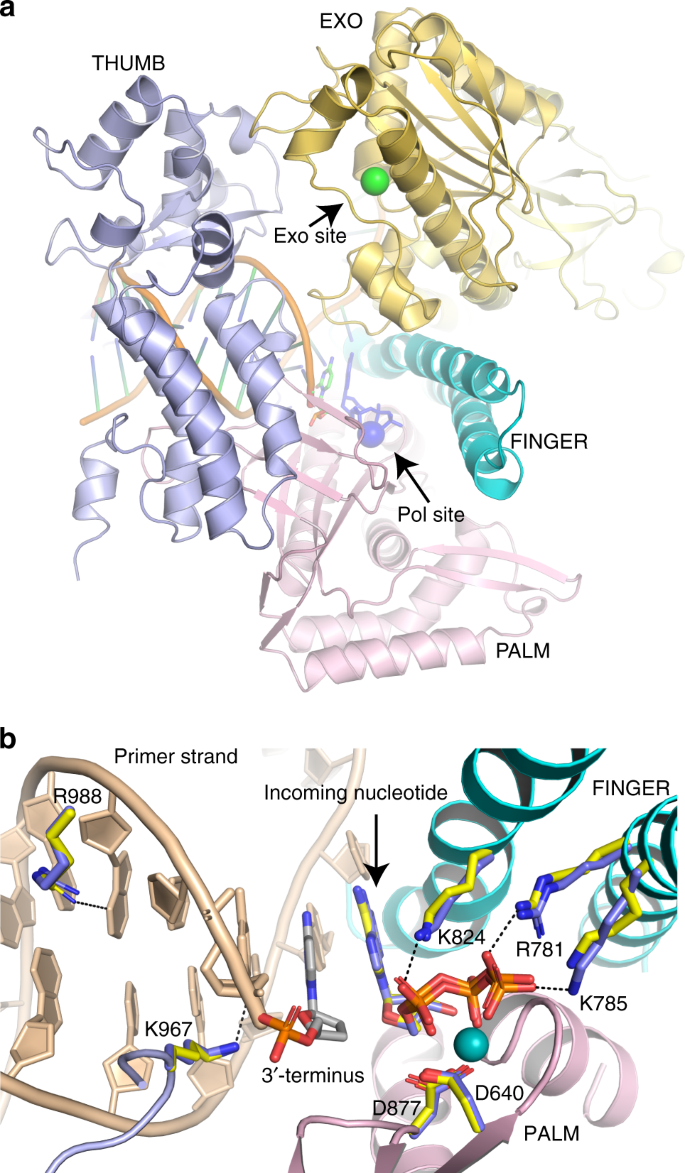
Structural consequence of the most frequently recurring cancer-associated substitution in DNA polymerase ε | Nature Communications

Alternative DNA secondary structure formation affects RNA polymerase II promoter-proximal pausing in human | Genome Biology | Full Text

Requirement for transient metal ions revealed through computational analysis for DNA polymerase going in reverse | PNAS

Integrator is recruited to promoter‐proximally paused RNA Pol II to generate Caenorhabditis elegans piRNA precursors | The EMBO Journal

Structure and function of the pol II transcription machinery | Taatjes Lab | University of Colorado Boulder

A fidelity mechanism in DNA polymerase lambda promotes error‐free bypass of 8‐oxo‐dG | The EMBO Journal
Mammalian Base Excision Repair: Functional Partnership between PARP-1 and APE1 in AP-Site Repair | PLOS ONE
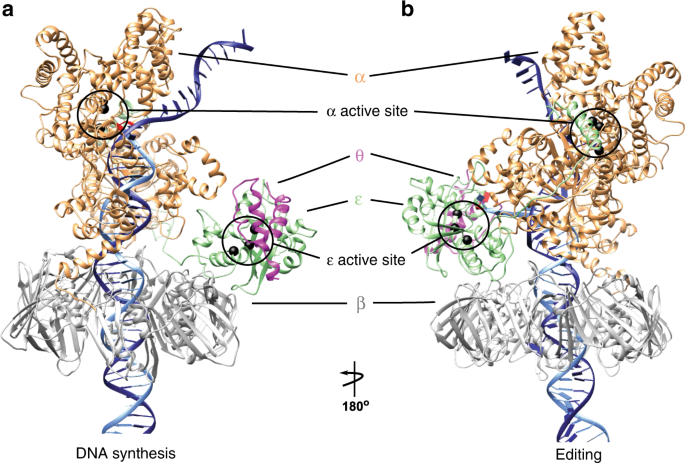
Polymerization and editing modes of a high-fidelity DNA polymerase are linked by a well-defined path | Nature Communications